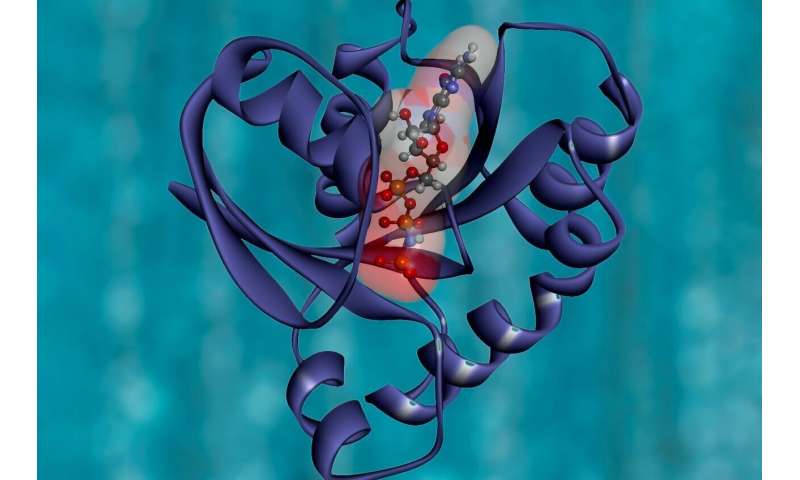
A pair of researchers on the University of California, San Francisco, has developed a brand fresh protein construction that allows for simplifying the strategy of custom-designing proteins. In their paper published within the journal Science, Nicholas Polizzi and William DeGrado discuss about their structural unit and the diagram in which they stale it. Anna Peacock, with the University of Birmingham, has published a Point of view piece outlining the work by the group in California within the identical journal challenge.
One among the things that chemists are requested to withhold out is custom manufacture proteins to be used in particular special functions. As the researchers display, doing so is thought about to be very disturbing. It customarily involves an excessive amount of trial and mistake which customarily translates to excessive model costs. In this fresh effort, the researchers have devised a brand fresh unit of protein construction to aid with such initiatives. They call it a van der Mer construction and record how it’d be stale to as we exclaim scheme ligand chemical group functionality to peptide residue backbone coordinates.
To near up with the fresh construction, the research pair poured thru and analyzed thousands of protein constructions within the Protein Data Bank. Their diagram differed from the norm in that they no longer popular the positioning of facet chains for the amino acid residue and as an different centered on the chemical teams that contacted the residues. The usage of this draw, they had been in a position to manufacture two fresh proteins that will be stale to name a drug called apixaban. Notably, their diagram enthusiastic making proper six sequences. Before their work, this sort of process would have in overall taken many more.
The name for the fresh construction came from combining van der Waals gorgeous forces with rotamers—the forms of sidechain conformations that amino acids can take hold of. The fresh construction works by mapping the backbones of amino acids to areas of chemical compounds within the Protein Data Bank obsessed on interactions with them. The researchers display that top seemingly no longer too long ago has the knowledge monetary institution when it comes to withhold ample files to allow for its exhaust in such an software. And they also display that the methodology and construction will also be stale to manufacture supply vehicles in step with proteins and likewise diminutive molecule functions.
More files:
Nicholas F. Polizzi et al. An outlined structural unit enables de novo manufacture of diminutive-molecule–binding proteins, Science (2020). DOI: 10.1126/science.abb8330
© 2020 Science X Network
Citation:
New structural unit simplifies the strategy of custom-designing proteins (2020, September 8)
retrieved 8 September 2020
from https://phys.org/news/2020-09-custom-designing-proteins.html
This doc is topic to copyright. Rather than any beautiful dealing for the reason of inner most gape or research, no
half is susceptible to be reproduced with out the written permission. The voice material is geared up for files functions top seemingly.



