
Chromosomes search for various than you suspect
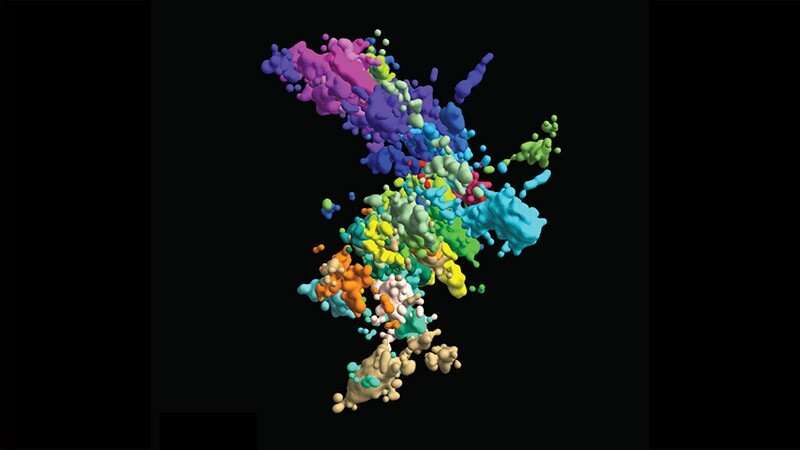
In high faculty textbooks, human chromosomes are pictured as wonky Xs devour two hotdogs jammed together. But those photos are a ways from correct. “For 90 p.c of the time,” acknowledged Jun-Han Su, “chromosomes originate now not exist devour that.”
Final year, sooner than Su graduated alongside with his Ph.D., he and three most up-to-date Ph.D. candidates within the Graduate College of Arts and Sciences—Pu Zheng, Seon Kinrot and Bogdan Bintu—captured high-resolution 3-d photos of human chromosomes, the complicated houses for our DNA. Now, those photos can also present enough proof to trade those Xs into extra complicated nonetheless a ways extra correct symbols to now not most inviting deliver the following abilities of scientists nonetheless back the most up-to-date abilities unravel mysteries about how chromosome structure influences feature.
All living issues, folks included, need to procure current cells to interchange those too extinct and extinct-out to feature. To achieve that, cells divide and replicate their DNA, which is wrapped into labyrinthine libraries interior chromatin, the stuff interior chromosomes. Extended in a straight line, DNA in a single cell can reach six toes, all of which will get wrapped into tight, complicated constructions in a cell nucleus. Honest one mistake copying or re-winding that genetic materials can also field off genes to mutate or malfunction.
Zooming in shut enough to discover chromatin structure is great. But having a search for at each structure and own is extra great peaceful. Now, in a paper published in August in Cell, Zhuang and her group document a brand current methodology to image the structure and habits of chromatin together, connecting the dots to resolve how one influences the different to retain acceptable feature or field off disease.
“It’s somewhat crucial to resolve the 3-d organization,” acknowledged Zhuang, the David B. Arnold, Jr. Professor of Science, “to imprint the molecular mechanisms underlying the organization and to also imprint how this organization regulates genome feature.”
With their current high-resolution 3-d imaging methodology, the group started to originate a chromosomal map from each huge-lens photos of all 46 chromosomes and shut-u.s.of one a part of one chromosome. To image one thing that is peaceful too minute to image, they captured linked dots (“genomic loci”) alongside every DNA chain. By connecting a form of dots, they’ll also procure a complete image of the chromatin structure.
But there used to be a snag. Beforehand, Zhuang acknowledged, the sequence of dots they’ll also image and name used to be restricted by the sequence of colours they’ll also image together: three. Three dots can now not originate a complete image.
So, Zhuang and her group came up with a sequential methodology: Image three loads of loci, quench the signal, after which image every other three in immediate succession. With that methodology, every dot will get two figuring out marks: colour and image round.
“Now we even own 60 loci simultaneously imaged and localized and, importantly, identified,” acknowledged Zhuang.
Tranquil, to conceal the overall genome, they wanted extra—thousands—so they modified into to a language that is already inclined to field up and retailer good amounts of recordsdata: binary. By imprinting binary barcodes on loads of chromatin loci, they’ll also image a ways extra loci and decode their identities later. Shall we embrace, a molecule imaged in round one nonetheless now not round two will get a barcode initiating with “10.” With 20-bit barcodes, the group can also differentiate 2,000 molecules in factual 20 rounds of imaging. “In this combinatorial methodology, we can elevate the sequence of molecules which could well be imaged and identified critical extra suddenly,” acknowledged Zhuang.
With this methodology, the group imaged about 2,000 chromatin loci per cell, a extra than ten-fold elevate from their previous work and enough to procure a high-resolution image of what the structure of chromosomes looks devour in its native habitat. But they did not live there: They also imaged transcription issue (when RNA replicates genetic materials from DNA) and nuclear constructions devour nuclear speckles and nucleoli.
With their 3-d Google Maps of the genome, they’ll also launch to study how the structure shifts over time and how those territorial actions back or harm cell division and replication.
Researchers already know chromatin is broken into loads of areas and domains (devour deserts versus cities). But what those terrains search for devour in loads of cell types and how they feature is peaceful unknown. With their high-resolution photos, Zhuang and group definite that areas with many of genes (“gene-rich”) are inclined to flock to same areas on any chromosome. But areas with few genes (“gene-uncomfortable”) most inviting approach together if they part the same chromosome. One opinion is that gene-rich areas, which could well be active sites for gene transcription, approach together devour a factory to enable extra efficient manufacturing.
Whereas extra study is foremost sooner than confirming this opinion, one thing is now definite: local chromatin atmosphere impacts transcription issue. Structure does influence feature. The group also chanced on that no two chromosomes search for a connected, even in cells which could well be otherwise same. To search for what every chromosome looks devour in every cell within the human body will resolve a ways extra work than one lab can resolve on on my own.
“It’s miles now not going to be capability to originate factual on our work,” Zhuang acknowledged. “We want to originate on many, many labs’ work in tell to own a complete working out.”
More recordsdata:
Jun-Han Su et al, Genome-Scale Imaging of the 3D Organization and Transcriptional Screech of Chromatin, Cell (2020). DOI: 10.1016/j.cell.2020.07.032
Journal recordsdata:
Cell
Quotation:
Chromosomes search for various than you suspect (2020, November 18)
retrieved 19 November 2020
from https://phys.org/news/2020-11-chromosomes.html
This tale is self-discipline to copyright. Other than any fair dealing for the rationale of deepest detect or study, no
part would be reproduced without the written permission. The train is equipped for recordsdata purposes most inviting.