Oncotarget: Genome extensive DNA methylation panorama finds glioblastoma’s impact

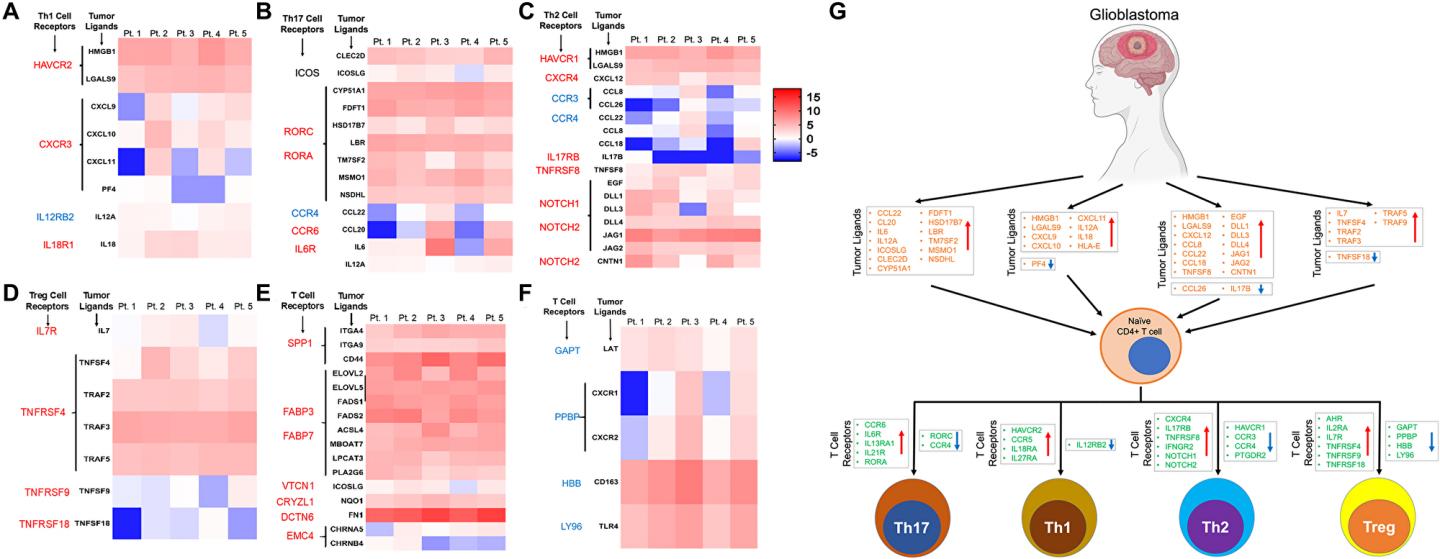
IMAGE: Expression pattern of ligands of corresponding receptors specific for CD4+ T cell lineages in GBM tumor tissue. (A-D) The heatmaps account for expression of the ligands in tumor tissue and graphical…
ogle more
Credit rating: Correspondence to – Mahua Dey – [email protected]
Oncotarget printed “Genome extensive DNA methylation panorama finds glioblastoma’s impact on epigenetic changes in tumor infiltrating CD4+ T cells” which reported that entire-genome bisulfite sequencing of tumor infiltrating and blood CD4 T-cell from GBM patients showed 13571 differentially methylated regions and a obvious methylation pattern of methylation of tumor infiltrating CD4 T-cells with necessary inter-affected person variability.
The methylation changes also resulted in transcriptomic changes with 341 differentially expressed genes in CD4 tumor infiltrating T-cells when in contrast to blood.
Analysis of specific genes smitten by CD4 differentiation and efficiency printed differential methylation space of TBX21, GATA3, RORC, FOXP3, IL10 and IFNG in tumor CD4 T-cells.
Interestingly, the authors seen dysregulation of several ligands of T cell plot genes in GBM tissue corresponding to the T-cell receptors that were dysregulated in tumor infiltrating CD4 T-cells.
These Oncotarget outcomes point out that GBM might well per chance induce epigenetic alterations in tumor infiltrating CD4 T-cells there by influencing anti-tumor immune response by manipulating differentiation and efficiency of tumor infiltrating CD4 T-cells.
These Oncotarget outcomes point out that GBM might well per chance induce epigenetic alterations in tumor infiltrating CD4 T-cells
Dr. Mahua Dey from The University of Wisconsin-Madison moreover to The Indiana University School of Medication stated, “Naïve CD4+ helper T cell inhabitants is famous for its polyfunctionality and extremely plastic characteristics.“
In the tumor microenvironment, lineage commitments of CD4 T cells replicate initiation of recent applications of gene expression inner tumor infiltrating naïve T cells.
The GBM tumor microenvironment is famous to be extremely immunosuppressive, possessing just a few outlandish properties including:
- Impaired cellular immunity no dearth of tumor infiltrating T cells
- Excessive ranges of TGFβ secreted by resident moreover to circulating microglia and
- Expression of several inhibitory ligands, eliciting anergy and apoptosis of cytotoxic lymphocytes in the TME, immune checkpoints expression, and increased infiltration of immunosuppressive cells.
Genome extensive methylation sequencing showed 13571 uniquely differentially methylated regions , mostly concentrated around the TSS, in the CD4 T cells from GBM affected person tumor when in contrast to blood.
Moreover, combining transcriptomic recordsdata from RNAseq diagnosis with DNA methylation, we seen differential methylation of gene devices specific for CD4 T cells including Th1, Th2, Th17 and iTregs in GBM tumors, despite the truth that with necessary interpatient variability.
In conclusion, this recordsdata for the critical time, narrative outlandish DNA methylation pattern and gene expression profiles in GBM associated tumor infiltrating CD4 T cells when in contrast to CD4 T-cell from the blood of the equivalent affected person and a few of their ligands on the GBM cells suggesting that CD4 T cells plot and differentiation is inclined to be influenced by the GBM TME by system of epigenetic mechanisms comparable to, DNA methylation.
The Dey Be taught Crew concluded of their Oncotarget Be taught Output, “in the unique scientific corelative narrative, we demonstrated that differential DNA methylation pattern might well per chance impact gene expression in tumor infiltering CD4+ T cells as when in contrast to circulating blood CD4+ T cells in GBM patients. Our findings present evidence that GBM will possible be influencing the tell of tumor infiltrating CD4+ T cells by epigenetic modification in the develop of DNA methylation of key immune plot regulating genes and influencing the fate of helper T cells in the GBM TME. In line with our observations we deem that perchance epigenetic interaction between GBM and tumor infiltrating CD4+ T cells is responsible for the immunosuppressed tell viewed in the GBM patients. Our recordsdata convincingly account for that there is necessary inter-affected person variability in the GBM tumor ligand expression of varied T-cell modulating ligands and in consequence placing differences in the methylation pattern and gene expression in tumor infiltrating CD4+ T-cells. This has a really solid implication for deciding on future patients for immunotherapy trials who might well per chance have better probability of responding to immunotherapy than others in step with their tumor immune signature. The findings from our corelative hit upon wants to be extra validated in the experimental atmosphere.“
###
DOI – https:/
Elephantine text – https:/
Correspondence to – Mahua Dey – [email protected]
Key phrases –
glioblastoma,
malignant glioma,
CD4+ T cell,
DNA methylation,
mind cancer
About Oncotarget
Oncotarget is a bi-weekly, hit upon-reviewed, birth gather admission to biomedical journal overlaying evaluate on all parts of oncology.
To be taught more about Oncotarget, please search the suggestion of with https:/
SoundCloud – https:/
Fb – https:/
Twitter – https:/
LinkedIn – https:/
Pinterest – https:/
Reddit – https:/
Oncotarget is printed by Affect Journals, LLC please search the suggestion of with https:/
Disclaimer: AAAS and EurekAlert! are no longer responsible for the accuracy of recordsdata releases posted to EurekAlert! by contributing institutions or for the utilization of any recordsdata throughout the EurekAlert design.