Pulling the slide on the coronavirus reproduction machine
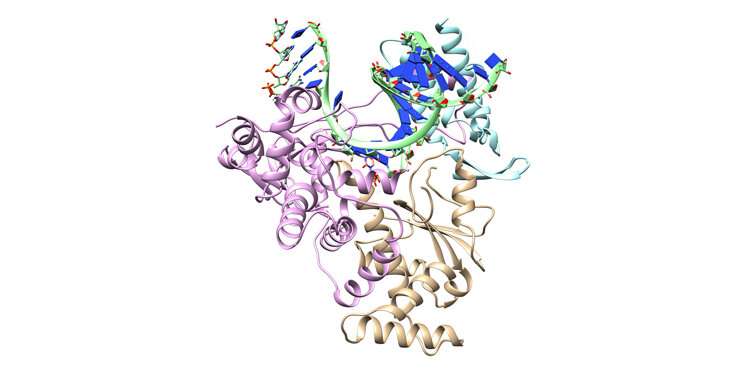
Key proteins outdated by coronavirus for its reproduction being modeled on NSF-funded Frontera supercomputer by Andres Cisneros examine neighborhood of the University of North Texas. Learn targets encompass finding ways to enhance on COVID-19 therapeutic remdesivir. NSF-funded Frontera allocation awarded to Cisneros thru the COVID-19 High Efficiency Computing Consortium.
In Can even merely 2020, the U.S. Meals and Drug Administration approved the antiviral drug remdesivir for emergency treatment of COVID-19, one of easiest four therapeutics in the in the intervening time with this case. Remdesivir stops the chemical equipment that the coronavirus makes employ of to reproduction itself, binding to an enzyme that does the assembly. Whereas remdesivir has confirmed promise in helping patients salvage successfully from COVID-19, scientists are investigating ways to enhance its effectiveness.
A crew of scientists led by G. Andres Cisneros of the University of North Texas is modeling the essential ingredients of the coronavirus that it makes employ of to reproduction itself. The simulations are being done on the Stampede2 and Frontera supercomputers on the Texas Evolved Computing Heart (TACC).
“We were very fortunate to be granted an allocation on Frontera to be ready to work on investigating the mechanism of gear that take mark to 2 particular proteins in COVID-19,” Cisneros said. His work investigates how remdesivir and various available capsules inhibit the proteins NSP-12 and the essential protease, each enzymes the coronavirus needs for replication. “By how these capsules attain their work, perchance this records will also be outdated to enhance upon them.”
The NSP-12 protein locations together the nucleotides that affect up viral RNA, abbreviated as A, U, G, and C, constructing total sets of genetic material for contemporary coronavirus copies. NSP-12 is in actuality phase of a greater construction known as the RNA-dependent RNA polymerase (RDRP) that copies the total RNA. Remdesivir binds with RDRP, plugging up the equipment.
“We’re investigating how this process happens,” said Cisneros. “By doing this, perchance there will possible be a blueprint for us and various scientists to come aid up with ideas on whether and how remdesivir will also be improved.”
The more than just a few protein Cisneros is studying is named the essential protease. It separates a polyprotein produced by SARS-CoV-2 translated from viral RNA into purposeful proteins that keep ‘meat’ on its viral bones. Quit the protease, and likewise you end the virus from forming. This makes it a mountainous drug goal.
Cisneros defined that he makes employ of the predominant math and physics of Newton’s equations and quantum mechanics to calculate the properties of the proteins, including all the pieces connected to its functioning, such because the RNA and water. An technique known as classical molecular dynamics makes employ of Newton’s equations to simulate how the proteins transfer and have interaction dynamically in time. “We’re talking about methods that we simulate that are in the a complete bunch of thousands of atoms,” Cisneros said.
He furthermore simulates the chemical reactions contained in the proteins to research how the capsules end RDRP or the protease. A hybrid potential known as QM/MM (quantum mechanics/molecular mechanics) saves computational time and money by focusing more closely on interactions on the active blueprint, the utilization of the more approximate straight molecular dynamics for all the pieces else.
The Cisneros neighborhood developed and maintains a program known as LICHEM that lets them employ the QM/MM technique. “One among the aspects of LICHEM is that it permits us to employ approaches for the classical mechanics phase that encompass a greater description of the physics that are taking place between the molecules in the classical atmosphere, namely, the AMOEBA attainable” Cisneros said. AMOEBA is developed by Pengyu Ren of UT Austin; Jay Ponder of the University of Washington; and Jean-Philip Piquemal at Sorbonne University in Paris with contributions from the Cisneros neighborhood for ionic liquids.
“Frontera, with now not easiest compute energy however the intercommunication between the nodes, permits us to urge these QM/MM calculations with distinguished greater, now not easiest velocity, however furthermore throughput,” Cisneros said. Frontera freed them to urge multiple methods at a time. “In my neighborhood, I even have 5 various scientists, graduate students and publish docs, that are engaged on each of these methods, however in various pieces of the puzzle. All of them are ready to entry these sources. Or now not it is positively very helpful, and we very distinguished take care of the allocation.”
What received Cisneros going used to be news in April of 2020 of the crystal construction of the SARS-CoV-2 RDRP being reported. “I contacted my neighborhood and suggested them that with this records, there is something we can attain to aid with the pandemic,” he said.
Within two days of the news, Cisneros efficiently proposed his examine on coronavirus drug targets to the COVID-19 High Efficiency Computing Consortium. Dozens of nationwide and global supercomputing facilities, industry, and organizations including TACC have volunteered their sources to the consortium in enhance of scientists’ effort to fight the coronavirus.
The allocation used to be on the foundation awarded merely on TACC’s Stampede2, the supercomputing flagship of the Nationwide Science Foundation (NSF) that is ranked 21st fastest in the realm and #2 for academic methods essentially based completely on the Top500. “Then we were contacted by TACC and grateful that we were granted entry to Frontera. Now now we have gotten entry to each methods, which is most continuously mountainous,” Cisneros said.
The Frontera supercomputer is the #1 fastest academic supercomputer and #8 fastest worldwide. Both Frontera and Stampede2 are funded by the NSF.
“We’re very joyful with this design. We were ready to transfer just a few of the records that we had from Stampede2 to Frontera,” Cisneros said. One among his now not too long prior to now graduated students, Erik Vazquez Montelongo, blueprint up the total calculations for LICHEM on Frontera essentially based completely on what he learned on Stampede2. “That no doubt has been a boon. Frontera for our calculations has been working in actuality successfully. We’re in actuality joyful with it.”
One among the postdocs in The Cisneros Group, Sehr Nazeem-Kahn, generated the model for RDRP, the remdesivir and various drug candidates, all in the active blueprint. With that in hand, they started working simulations.
“We were very joyful to glimpse that her model used to be in fact very shut to the experimental construction. That is in actuality helpful for us, since it validates the model that has been built by the neighborhood and shows that we are on the beautiful observe,” he added.
At this time, Dr. Naseem-Khan is working molecular dynamics simulations of this model with remdesivir on Frontera. “We are furthermore starting with our QM/MM calculations for RDRP. Within the case for the essential protease, there have been structures that furthermore major to be modeled and attributable to this fact were confirmed. That used to be furthermore very stress-free,” Cisneros said.
With that construction records, they’re six various inhibitor molecules. “A form of, we’re already starting QM/MM calculations on Frontera, and one other one on Stampede2,” Cisneros said. If all goes successfully, he’s hoping to salvage results in the next 5 to six months. “These are very costly calculations,” he added. “Furthermore, working the prognosis takes time. If we were to employ merely the sources at dwelling, it would bewitch plenty of years.”
Citation:
Pulling the slide on the coronavirus reproduction machine (2020, September 19)
retrieved 20 September 2020
from https://phys.org/news/2020-09-coronavirus-machine.html
This doc is subject to copyright. Other than any beautiful dealing for the reason of personal ogle or examine, no
phase will possible be reproduced with out the written permission. The vow material is equipped for records purposes easiest.